Portfolio item number 1
Short description of portfolio item number 1
Short description of portfolio item number 1
Short description of portfolio item number 2 
Published in Biomed Opt Express, 2012, Google Scholar Link
Published in J Vis Exp, 2013, Google Scholar Link
Published in Am J Respir Crit Care Med, 2013, Google Scholar Link
Published in Chest, 2013, Google Scholar Link
Published in Chest, 2013, Google Scholar Link
Published in Opt Express, 2013, Google Scholar Link
Published in Adv Funct Mater, 2014, Google Scholar Link
Published in Proc Natl Acad Sci U S A, 2015, Google Scholar Link
Published in ACS Biomater Sci Eng, 2015, Google Scholar Link
Published in Biomed Opt Express, 2015, Google Scholar Link
Published in Adv Mater, 2016, Google Scholar Link
Published in Applied Physics Letters, 2016, Google Scholar Link
Published in Biomaterials, 2016, Google Scholar Link
Published in ACS Biomater Sci Eng, 2016, Google Scholar Link
Published in ACS Omega, 2017, Google Scholar Link
Published in Soft Matter, 2017, Google Scholar Link
Published in Nat Nanotechnol, 2017, Google Scholar Link
Published in J Biomed Opt, 2017, Google Scholar Link
Published in Opt Lett, 2018, Google Scholar Link
Published in Biomed Opt Express, 2018, Google Scholar Link
Published in Clin Cancer Res, 2019, Google Scholar Link
Published in Respirology, 2019, Google Scholar Link
Published in , 1900, Google Scholar Link
Published in Journal of Biomedical Optics, 2020, Google Scholar Link
Published in APL Photonics, 2020, Google Scholar Link
Published in Biomed Opt Express, 2021, Google Scholar Link
Published in IEEE Trans Biomed Eng, 2021, Google Scholar Link
Published in Journal of Medical Imaging, 2023, Google Scholar Link
Worked on in: – (Publication link)
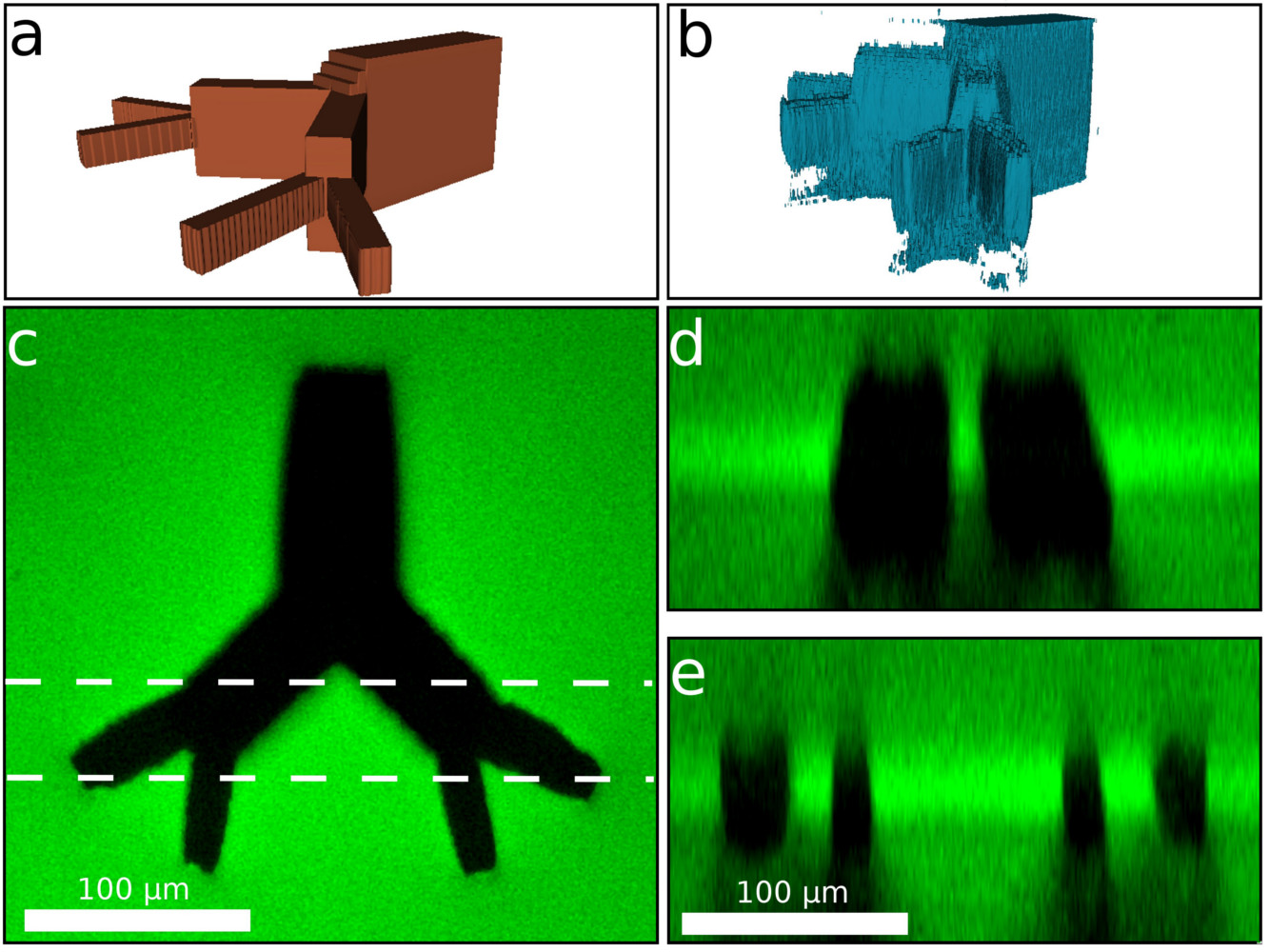
No cell in your body is more than a few hundreds of microns from a blood vessel. A key challenge for artificial organs is developing a biomimetic vasculature to support cell growth. In this work I used the large three-photon absorption cross section of silk fibroin to generate 3D structures in silk hydrogels that range in size from a few microns up to nearly a millimeter at unprecedented depths
Worked on in: – (Publication link)

Silk is a great biocompatible, transparent substrate for all sorts of biomedical applications. One such is to use silk to patch corneal wounds. I developed a method to transform a liquid silk solution into a hydrogel when exposed to blue or UV light that binds readily to corneal tissue. A provisional patent on this method has been filed (62/268,993).
Worked on in: – (Publication link)
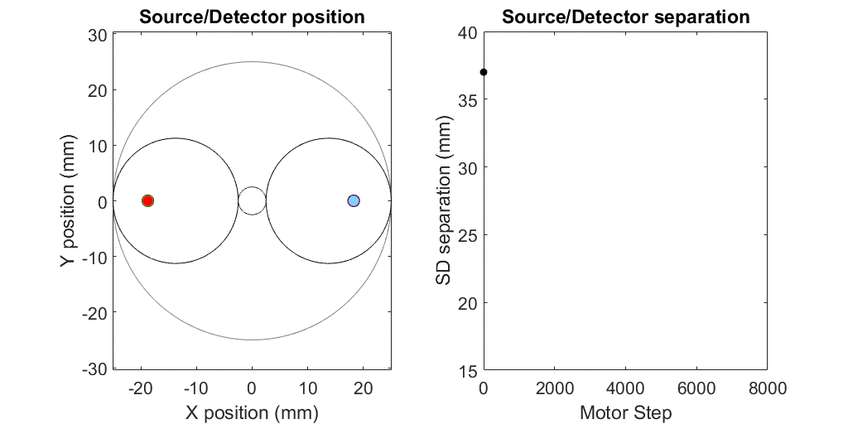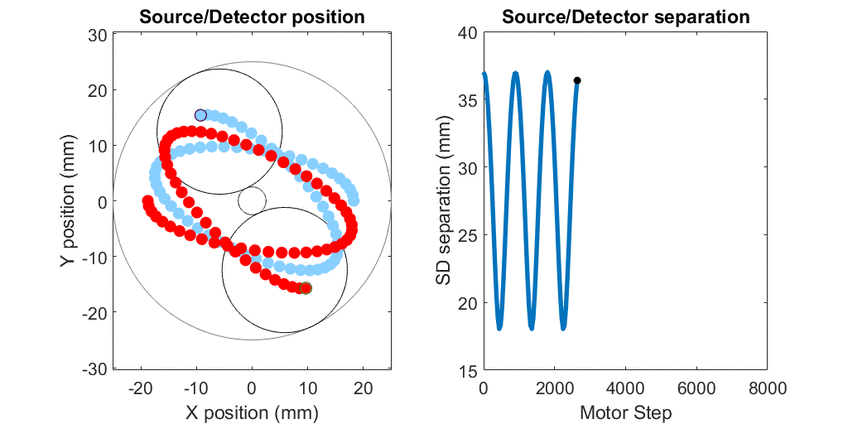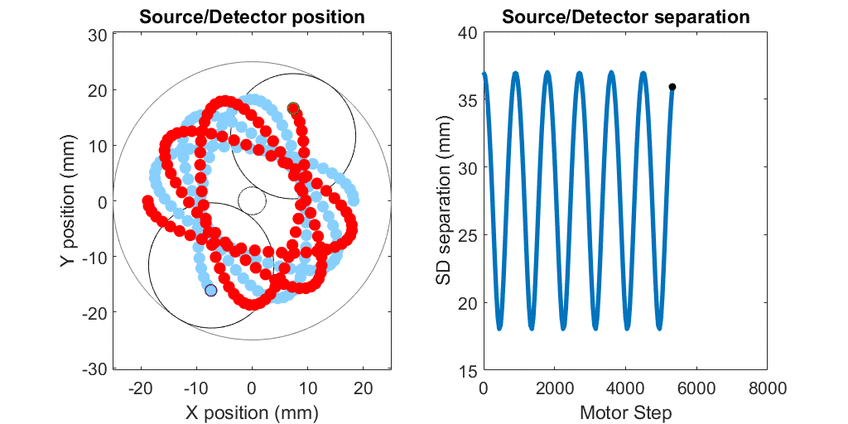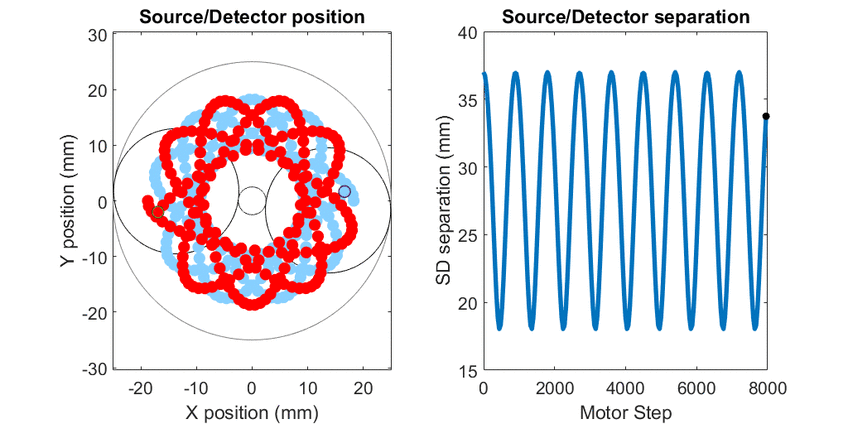
In diffuse optical spectroscopy, the distance between the source and detector fiber determines how deep below the surface detected light has traveled. Longer separation is more sensitive to deeper structures, while short separation is more sensitive to superficial changes. Many instruments rely on an array of different sources and detectors to build up 3D estimates of tissue hemoglobin. Here, I use mechanical scanning to trace the source and detector through a hypotrochoid which allows for a range of source/detector separations that are all centered at the same location. This allows measurements to form a pseudo axial line scan to build up 2- and 3-dimensional images of tissue chromophores.
Worked on in: – (Publication link)

Spatial Frequency Domain Imaging is a diffuse optical imaging method able to quantify the concentration fo hemoglobin in tissue. OpenSFDI is a wesite that provides step-by-step instructions for building an SFDI system.
Worked on in: – (Publication link)
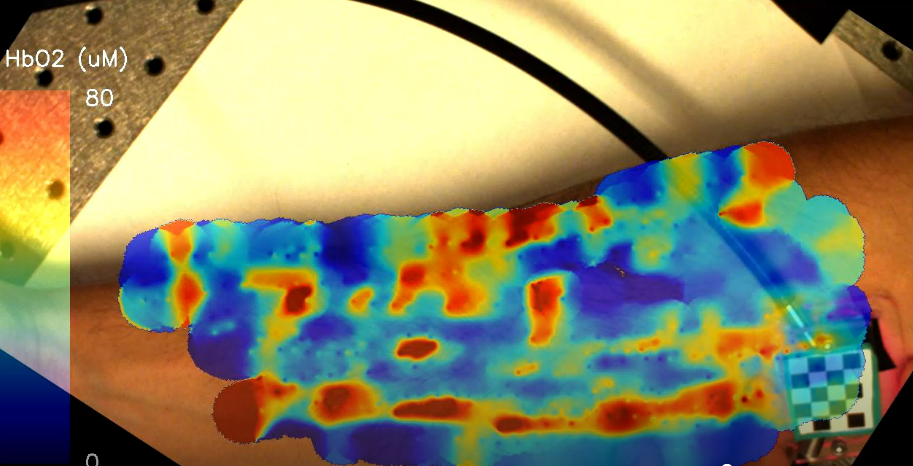
Diffuse optical spectroscopy has a long history of use for the monitoring of breast cancer, but lengthy acquisition and processing times have made it difficult to use in the clinic. While in the Roblyer lab, I helped develop an ultrafast spectroscopy system with onboard data processing and integrated it with a novel probe tracking technique to provide real-time information of tissue hemodynamics.
Worked on in:
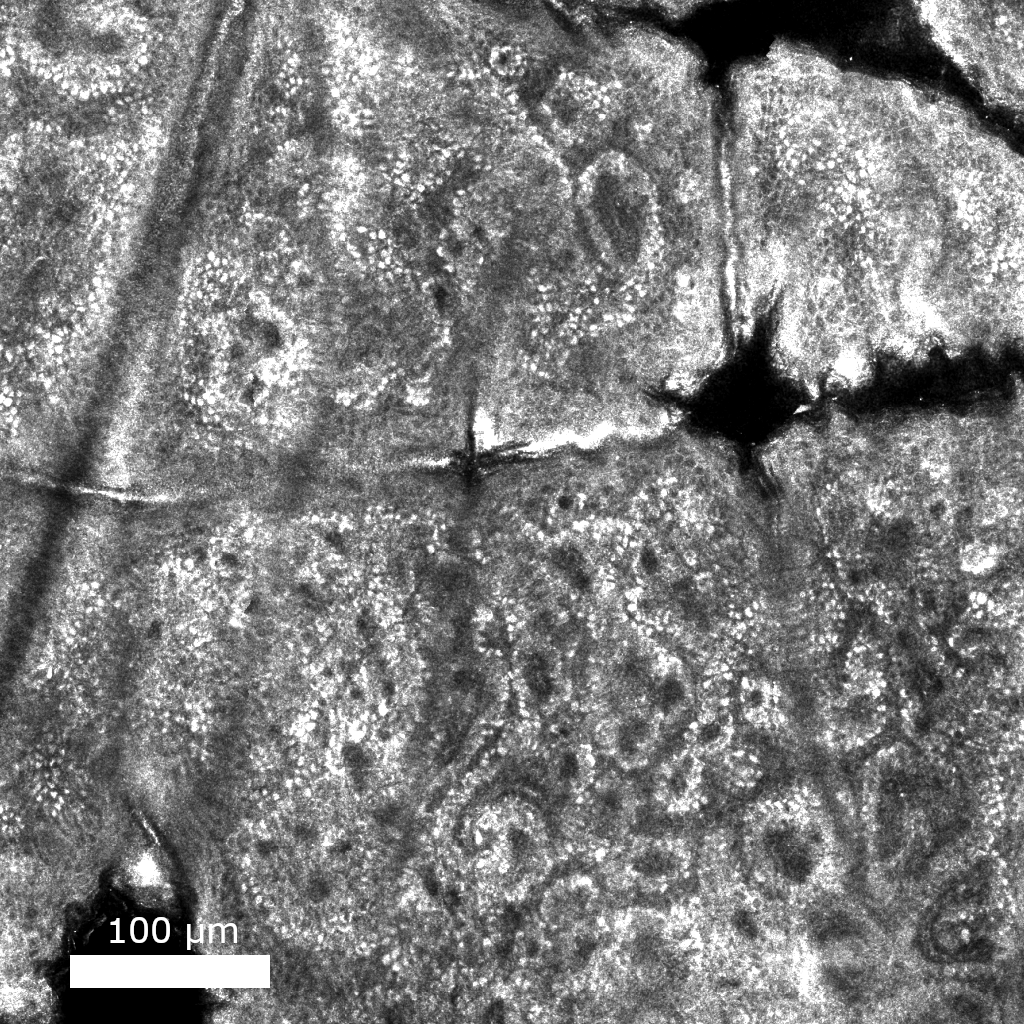
Reflectance confocal microscopy (RCM) is a noninvasive optical imaging modality capable of achieving cellular resolution that is currently used to rapidly diagnose skin cancer. I’m working on developing machine learning algorithms that will make it possible for the next generation of RCM microscopes smaller, faster, and cheaper.
Worked on in: – (Publication link)
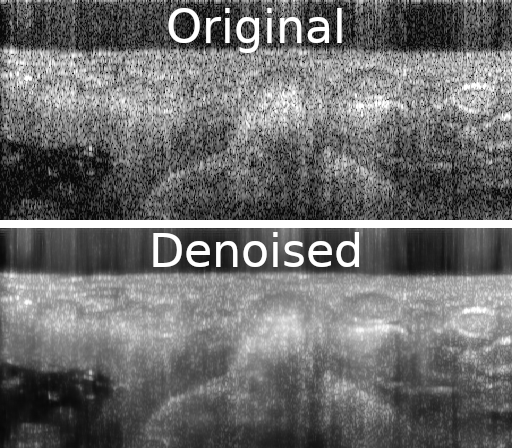
Improving medical images with machine learning is hard because of the difficulty in collecting datasets that contain gold standard, or ground truth images. Self-supervised methods can learn to denoise images solely from noisy data. In this work, I show how similarities between adjacent images in a volume can be used to remove noise from a variety of volumetric biomedical image data.
Published:
Published:
This presentation looks at optimal frequency selection for frequency-domain diffuse optical spectroscopy
Published:
This presentation described a novel instrument that allows users to scan over breast tissue and see maps of tissue oxygenation in real-time
Undergraduate course, University 1, Department, 2014
This is a description of a teaching experience. You can use markdown like any other post.
Workshop, University 1, Department, 2015
This is a description of a teaching experience. You can use markdown like any other post.