Self-supervised denoising of volumetric biomedical images
Worked on in: – (Publication link)
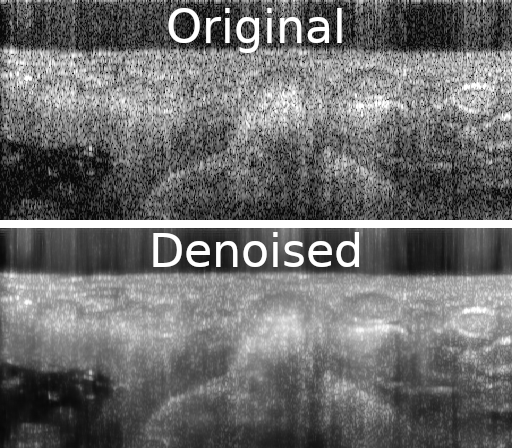
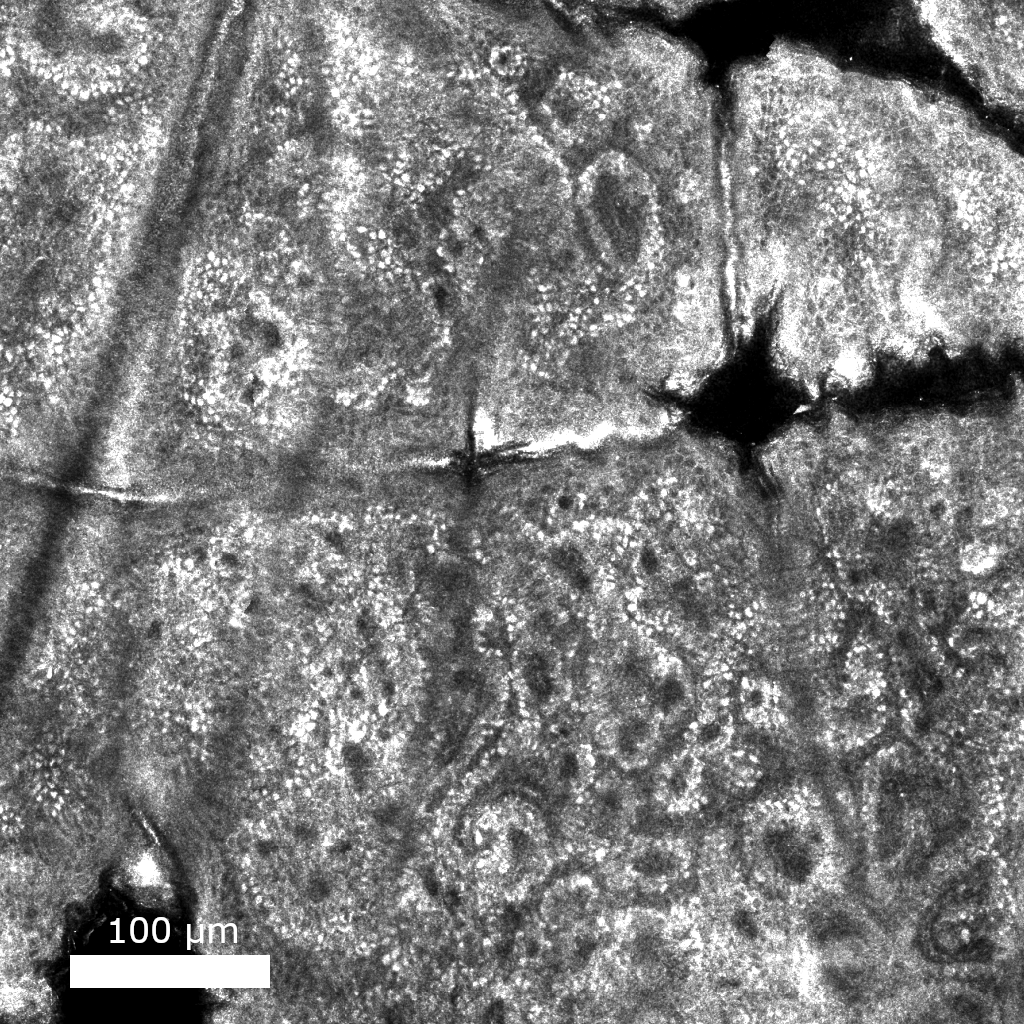
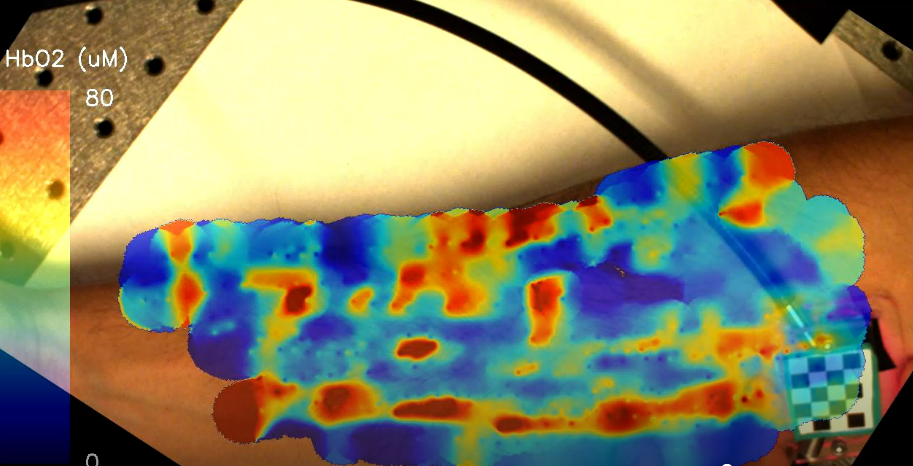
Improving medical images with machine learning is hard because of the difficulty in collecting datasets that contain gold standard, or ground truth images. Self-supervised methods can learn to denoise images solely from noisy data. In this work, I show how similarities between adjacent images in a volume can be used to remove noise from a variety of volumetric biomedical image data.